
Welcome to Watcher Forum
|
| | | How Coronavirus Can Be Stopped: 3D Atomic Map of COVID-19’s Viral Replication Mechanism |  |
| | | Author | Message |
|---|
Guest
Guest
 |  Subject: How Coronavirus Can Be Stopped: 3D Atomic Map of COVID-19’s Viral Replication Mechanism Subject: How Coronavirus Can Be Stopped: 3D Atomic Map of COVID-19’s Viral Replication Mechanism  Mon Nov 02, 2020 9:45 am Mon Nov 02, 2020 9:45 am | |
| https://scitechdaily.com/how-coronavirus-can-be-stopped-3d-atomic-map-of-covid-19s-viral-replication-mechanism/How Coronavirus Can Be Stopped: 3D Atomic Map of COVID-19’s Viral Replication MechanismTOPICS:BiochemistryCOVID-19DOEInfectious DiseasesMolecular PhysicsOak Ridge National LaboratoryPopularBy OAK RIDGE NATIONAL LABORATORY OCTOBER 31, 2020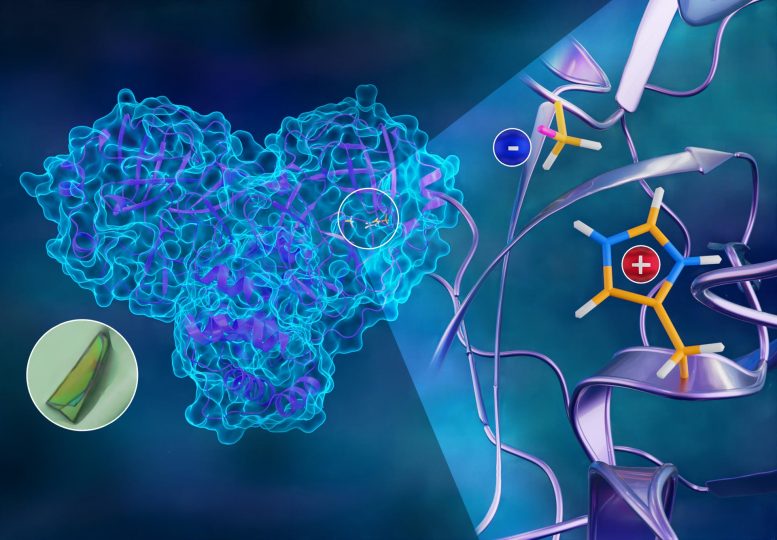
The first neutron structure of the SARS-CoV-2 main protease enzyme revealed unexpected electrical charges in the amino acids cysteine (negative) and histidine (positive), providing key data about the virus’s replication. Credit: Jill Hemman/ORNL, U.S. Dept. of EnergyTo better understand how the novel coronavirus behaves and how it can be stopped, scientists have completed a three-dimensional map that reveals the location of every atom in an enzyme molecule critical to SARS-CoV-2 reproduction.Researchers at the Department of Energy’s Oak Ridge National Laboratory used neutron scattering to identify key information to improve the effectiveness of drug inhibitors designed to block the virus’s replication mechanism. The research is published in the Journal of Biological Chemistry.The SARS-CoV-2 virus, which causes the COVID-19 disease, expresses long chains of proteins composed of approximately 1,900 amino acid residues. For the virus to reproduce, those chains have to be broken down and cut into smaller strands by an enzyme called the main protease. The active protease enzyme is formed from two identical protein molecules held together by hydrogen bonds. Developing a drug that inhibits or blocks the protease activity will prevent the virus from replicating and spreading to other cells in the body.“This new information is exactly what is needed to design inhibitors with a higher degree of specificity, ensuring the inhibitor molecules are binding very tightly to their intended targets and disabling the protease,” said ORNL’s Andrey Kovalevsky, corresponding author.Neutron experiments first revealed that the site containing the amino acids where the protein chains are cut is in an electrically charged reactive state and not in a resting or neutral state, contrary to previously held beliefs. Second, they mapped the location of each hydrogen atom in the places where inhibitors would bind to the protease enzyme, as well as the electrical charges of the associated amino acids. The experiments also charted the entire network of hydrogen bonds between the protein molecules that hold the enzyme together and enable it to initiate the chemical process of cutting the protein chains.“Half of the atoms in proteins are hydrogen. Those atoms are key players in enzymatic function and are essential to how drugs bind,” Kovalevsky said. “If we don’t know where those hydrogens are and how the electrical charges are distributed inside the protein, we can’t design effective inhibitors for the enzyme.”The team’s neutron study builds on previous research published in the journal Nature Communications, creating a complete atomic structure of the protease enzyme. The researchers have also made their data publicly available to the scientific community before both papers were published to accelerate solutions to the global pandemic.Neutrons are ideal probes for studying biological structures because they are nondestructive and highly sensitive to light elements such as hydrogen. The neutron scattering experiments were performed at the High Flux Isotope Reactor and the Spallation Neutron Source at ORNL. The protein samples were synthesized in adjacent facilities at the Center for Structural Molecular Biology.“This might be the quickest neutron structure of a protein ever produced. We started neutron experiments in May, and within five months, we obtained and published our results. That’s something that usually takes years,” said ORNL corresponding author Leighton Coates. “This work demonstrates what we can do at Oak Ridge. Everything was done here from start to finish. The proteins were expressed, purified, and crystallized, and all the data was collected and analyzed on site — a completely vertically integrated approach.”The team will now use the newly obtained information to investigate the binding properties of drug molecule candidates to produce improved COVID-19 therapeutics.“Not only is this the first time anyone has obtained a neutron structure of a coronavirus protein, but it’s also the first time anyone has looked at this class of protease enzymes using neutrons,” said ORNL’s Daniel Kneller, the study’s first author. “It’s an outstanding example of neutron crystallography serving the community when it needs it the most.”References:“Unusual zwitterionic catalytic site of SARS-CoV-2 main protease revealed by neutron crystallography” by Daniel W. Kneller, Gwyndalyn Phillips, Kevin L Weiss, Swati Pant, Qiu Zhang, Hugh M O’Neill, Leighton Coates and Andrey Kovalevsky, 15 October 2020, Journal of Biological Chemistry.
DOI: 10.1074/jbc.AC120.016154“Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography” by Daniel W. Kneller, Gwyndalyn Phillips, Hugh M. O’Neill, Robert Jedrzejczak, Lucy Stols, Paul Langan, Andrzej Joachimiak, Leighton Coates and Andrey Kovalevsky, 24 June 2020, Nature Communications.
DOI: 10.1038/s41467-020-16954-7In addition to Kneller, Coates, and Kovalevsky, the paper’s coauthors include Gwyndalyn Phillips, Kevin L. Weiss, Swati Pant, Qiu Zhang and Hugh M. O’Neill. COVID-19 research at ORNL is supported in part by the DOE Office of Science through the National Virtual Biotechnology Laboratory, a consortium of DOE national laboratories focused on response to COVID-19, with funding provided by the Coronavirus CARES Act.HFIR and SNS are DOE Office of Science User Facilities. |
|   | | Guest
Guest
 |  Subject: It’s Evolving: Coronavirus Genetic Mutation May Have Made COVID-19 More Contagious Subject: It’s Evolving: Coronavirus Genetic Mutation May Have Made COVID-19 More Contagious  Mon Nov 02, 2020 9:49 am Mon Nov 02, 2020 9:49 am | |
| https://scitechdaily.com/its-evolving-coronavirus-genetic-mutation-may-have-made-covid-19-more-contagious/It’s Evolving: Coronavirus Genetic Mutation May Have Made COVID-19 More ContagiousTOPICS:COVID-19EvolutionGeneticsPublic HealthUniversity Of Texas At AustinVirologyBy UNIVERSITY OF TEXAS AT AUSTIN NOVEMBER 1, 2020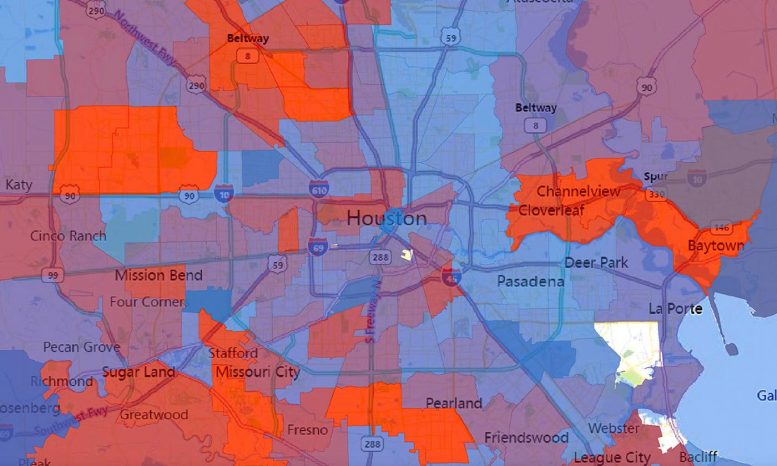 The number of virus strains present in each zip code in Houston during the second wave of COVID-19 cases in summer 2020. Number of strains is represented by a spectrum of colors from blue (0 strains) to red (50 strains). Credit: Houston Methodist/University of Texas at Austin. Credit: Houston Methodist/University of Texas at AustinA study involving more than 5,000 COVID-19 patients in Houston finds that the virus that causes the disease is accumulating genetic mutations, one of which may have made it more contagious. According to the paper published in the peer-reviewed journal mBIO, that mutation, called D614G, is located in the spike protein that pries open our cells for viral entry. It’s the largest peer-reviewed study of SARS-CoV-2 genome sequences in one metropolitan region of the U.S. to date.The paper shows “the virus is mutating due to a combination of neutral drift — which just means random genetic changes that don’t help or hurt the virus — and pressure from our immune systems,” said Ilya Finkelstein, associate professor of molecular biosciences at The University of Texas at Austin and co-author of the study. The study was carried out by scientists at Houston Methodist Hospital, UT Austin and elsewhere.During the initial wave of the pandemic, 71% of the novel coronaviruses identified in patients in Houston had this mutation. When the second wave of the outbreak hit Houston during the summer, this variant had leaped to 99.9% prevalence. This mirrors a trend observed around the world. A study published in July based on more than 28,000 genome sequences found that variants carrying the D614G mutation became the globally dominant form of SARS-CoV-2 in about a month. SARS-CoV-2 is the coronavirus that causes COVID-19.So why did strains containing this mutation outcompete those that didn’t have it?Perhaps they’re more contagious. A study of more than 25,000 genome sequences in the U.K. found that viruses with the mutation tended to transmit slightly faster than those without it and caused larger clusters of infections. Natural selection would favor strains of the virus that transmit more easily. But not all scientists are convinced. Some have suggested another explanation, called “founder’s effects.” In that scenario, the D614G mutation might have been more common in the first viruses to arrive in Europe and North America, essentially giving them a head start on other strains. The number of virus strains present in each zip code in Houston during the second wave of COVID-19 cases in summer 2020. Number of strains is represented by a spectrum of colors from blue (0 strains) to red (50 strains). Credit: Houston Methodist/University of Texas at Austin. Credit: Houston Methodist/University of Texas at AustinA study involving more than 5,000 COVID-19 patients in Houston finds that the virus that causes the disease is accumulating genetic mutations, one of which may have made it more contagious. According to the paper published in the peer-reviewed journal mBIO, that mutation, called D614G, is located in the spike protein that pries open our cells for viral entry. It’s the largest peer-reviewed study of SARS-CoV-2 genome sequences in one metropolitan region of the U.S. to date.The paper shows “the virus is mutating due to a combination of neutral drift — which just means random genetic changes that don’t help or hurt the virus — and pressure from our immune systems,” said Ilya Finkelstein, associate professor of molecular biosciences at The University of Texas at Austin and co-author of the study. The study was carried out by scientists at Houston Methodist Hospital, UT Austin and elsewhere.During the initial wave of the pandemic, 71% of the novel coronaviruses identified in patients in Houston had this mutation. When the second wave of the outbreak hit Houston during the summer, this variant had leaped to 99.9% prevalence. This mirrors a trend observed around the world. A study published in July based on more than 28,000 genome sequences found that variants carrying the D614G mutation became the globally dominant form of SARS-CoV-2 in about a month. SARS-CoV-2 is the coronavirus that causes COVID-19.So why did strains containing this mutation outcompete those that didn’t have it?Perhaps they’re more contagious. A study of more than 25,000 genome sequences in the U.K. found that viruses with the mutation tended to transmit slightly faster than those without it and caused larger clusters of infections. Natural selection would favor strains of the virus that transmit more easily. But not all scientists are convinced. Some have suggested another explanation, called “founder’s effects.” In that scenario, the D614G mutation might have been more common in the first viruses to arrive in Europe and North America, essentially giving them a head start on other strains.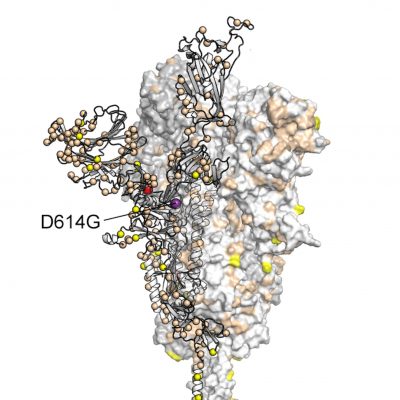
Most coronavirus strains circulating in Houston in the summer of 2020 had the D614G mutation in the spike protein. Credit: Houston Methodist/University of Texas at AustinThe spike protein is also continuing to accumulate additional mutations of unknown significance. The Houston Methodist-UT Austin team also showed in lab experiments that at least one such mutation allows spike to evade a neutralizing antibody that humans naturally produce to fight SARS-CoV-2 infections. This may allow that variant of the virus to more easily slip past our immune systems. Although it is not clear yet whether that translates into it also being more easily transmitted between individuals.The good news is that this mutation is rare and does not appear to make the disease more severe for infected patients. According to Finkelstein, the group did not see viruses that have learned to evade first-generation vaccines and therapeutic antibody formulations.“The virus continues to mutate as it rips through the world,” Finkelstein said. “Real-time surveillance efforts like our study will ensure that global vaccines and therapeutics are always one step ahead.”The scientists noted a total of 285 mutations across thousands of infections, although most don’t appear to have a significant effect on how severe the disease is. Ongoing studies are continuing to surveil the third wave of COVID-19 patients and to characterize how the virus is adapting to neutralizing antibodies that are produced by our immune systems. Each new infection is a roll of the dice, an additional chance to develop more dangerous mutations.“We have given this virus a lot of chances,” lead author James Musser of Houston Methodist told The Washington Post. “There is a huge population size out there right now.”Several other UT Austin authors contributed to the work: visiting scholar Jimmy Gollihar, associate professor of molecular biosciences Jason S. McLellan and graduate students Chia-Wei Chou, Kamyab Javanmardi and Hung-Che Kuo.The UT Austin team tested different genetic variants of the virus’s spike protein, the part that allows it to infect host cells, to measure the protein’s stability and to see how well it binds to a receptor on host cells and to neutralizing antibodies. Earlier in the year, McLellan and his team at UT Austin, in collaboration with researchers at the National Institutes of Health, developed the first 3D map of the coronavirus spike protein for an innovation that now factors into several leading vaccine candidates’ designs.The researchers found that SARS-CoV-2 was introduced to the Houston area many times, independently, from diverse geographic regions, with virus strains from Europe, Asia, South America and elsewhere in the United States. There was widespread community dissemination soon after COVID-19 cases were reported in Houston.Reference: “Molecular Architecture of Early Dissemination and Massive Second Wave of the SARS-CoV-2 Virus in a Major Metropolitan Area” by S. Wesley Long, Randall J. Olsen, Paul A. Christensen, David W. Bernard, James J. Davis, Maulik Shukla, Marcus Nguyen, Matthew Ojeda Saavedra, Prasanti Yerramilli, Layne Pruitt, Sishir Subedi, Hung-Che Kuo, Heather Hendrickson, Ghazaleh Eskandari, Hoang A. T. Nguyen, J. Hunter Long, Muthiah Kumaraswami, Jule Goike, Daniel Boutz, Jimmy Gollihar, Jason S. McLellan, Chia-Wei Chou, Kamyab Javanmardi, Ilya J. Finkelstein and James M. Musser, 30 October 2020, mBio.
DOI: 10.1128/mBio.02707-20This study was supported by the Fondren Foundation, Houston Methodist Hospital and Research Institute, the National Institutes of Health, the National Institute of Allergy and Infectious Diseases, the Welch Foundation, the National Science Foundation and the Defense Advanced Research Projects Agency. Ilya Finkelstein is a CPRIT scholar in cancer research, funded by the Cancer Prevention and Research Institute of Texas.An earlier version of the paper was posted in September to the preprint server medRxiv. |
|   | | | | How Coronavirus Can Be Stopped: 3D Atomic Map of COVID-19’s Viral Replication Mechanism |  |
|
Similar topics |  |
|
| | Permissions in this forum: | You cannot reply to topics in this forum
| |
| |
| |
|





